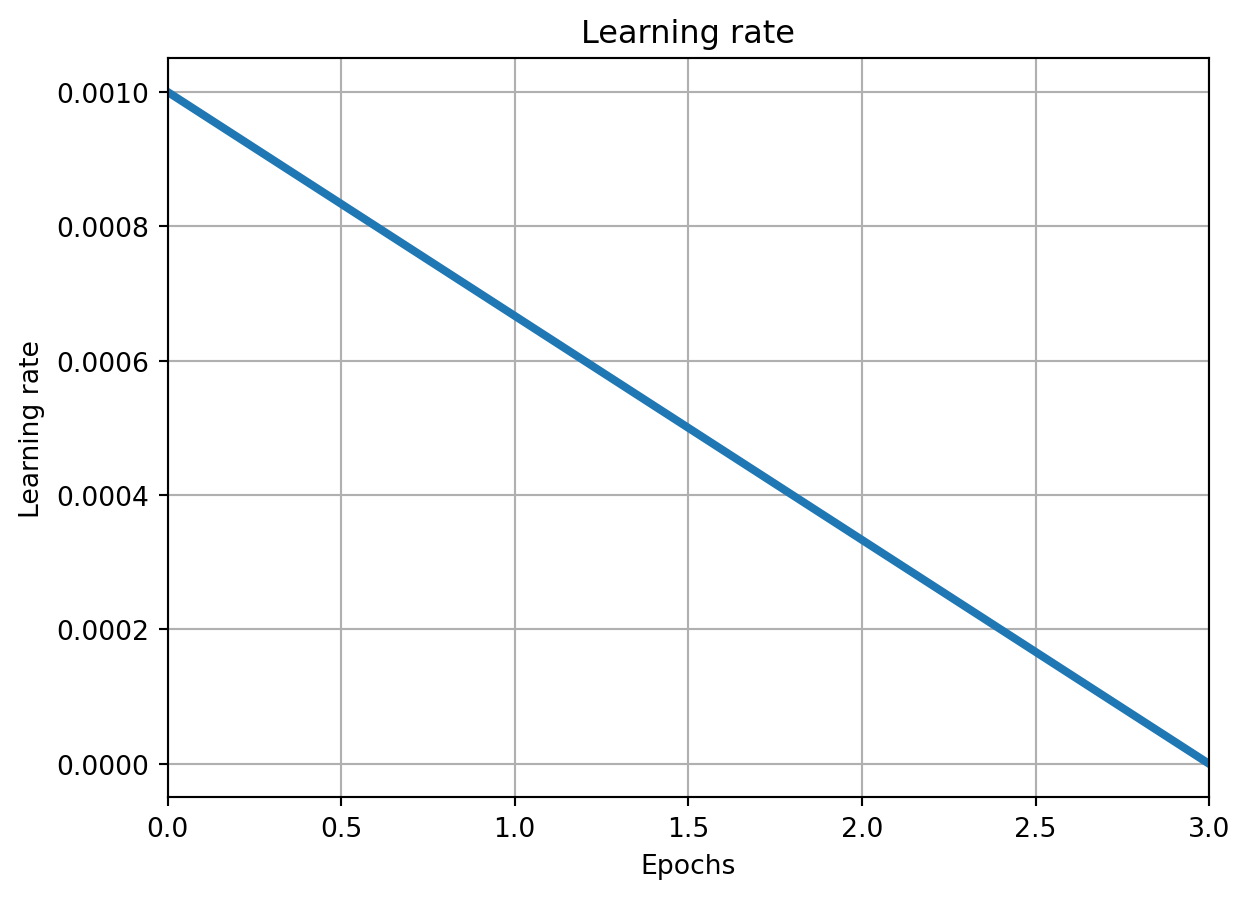
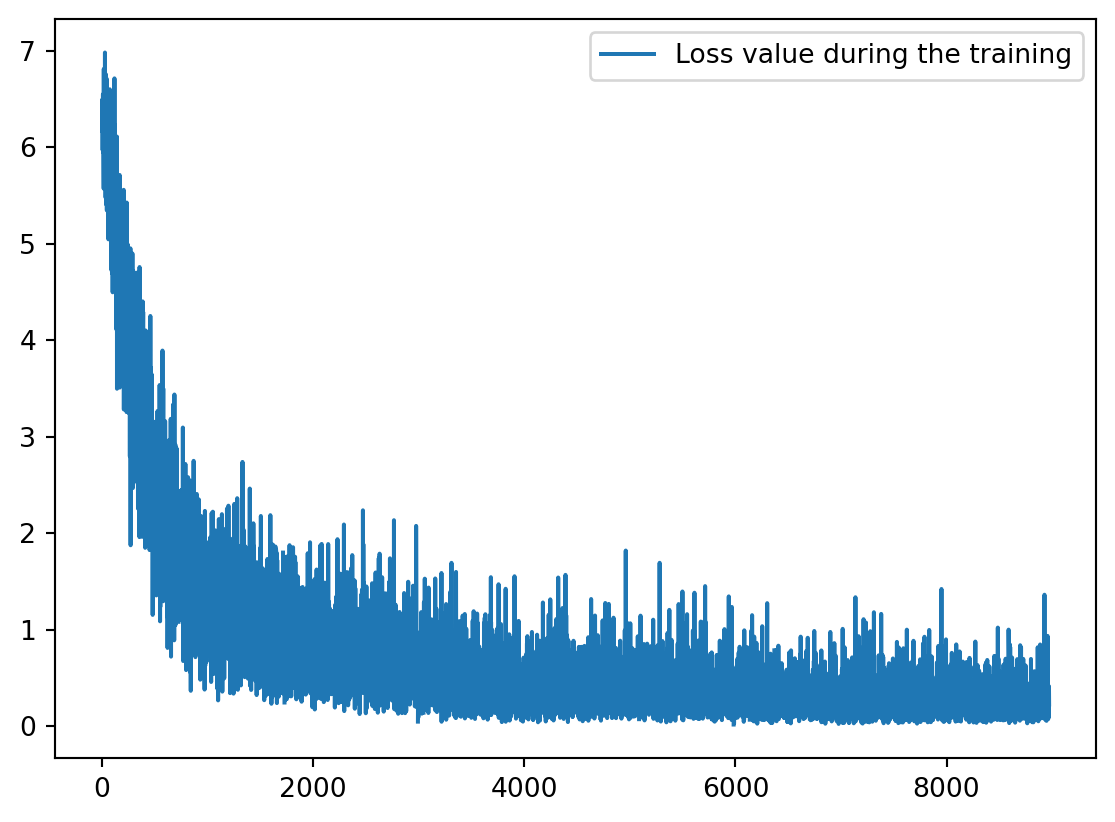
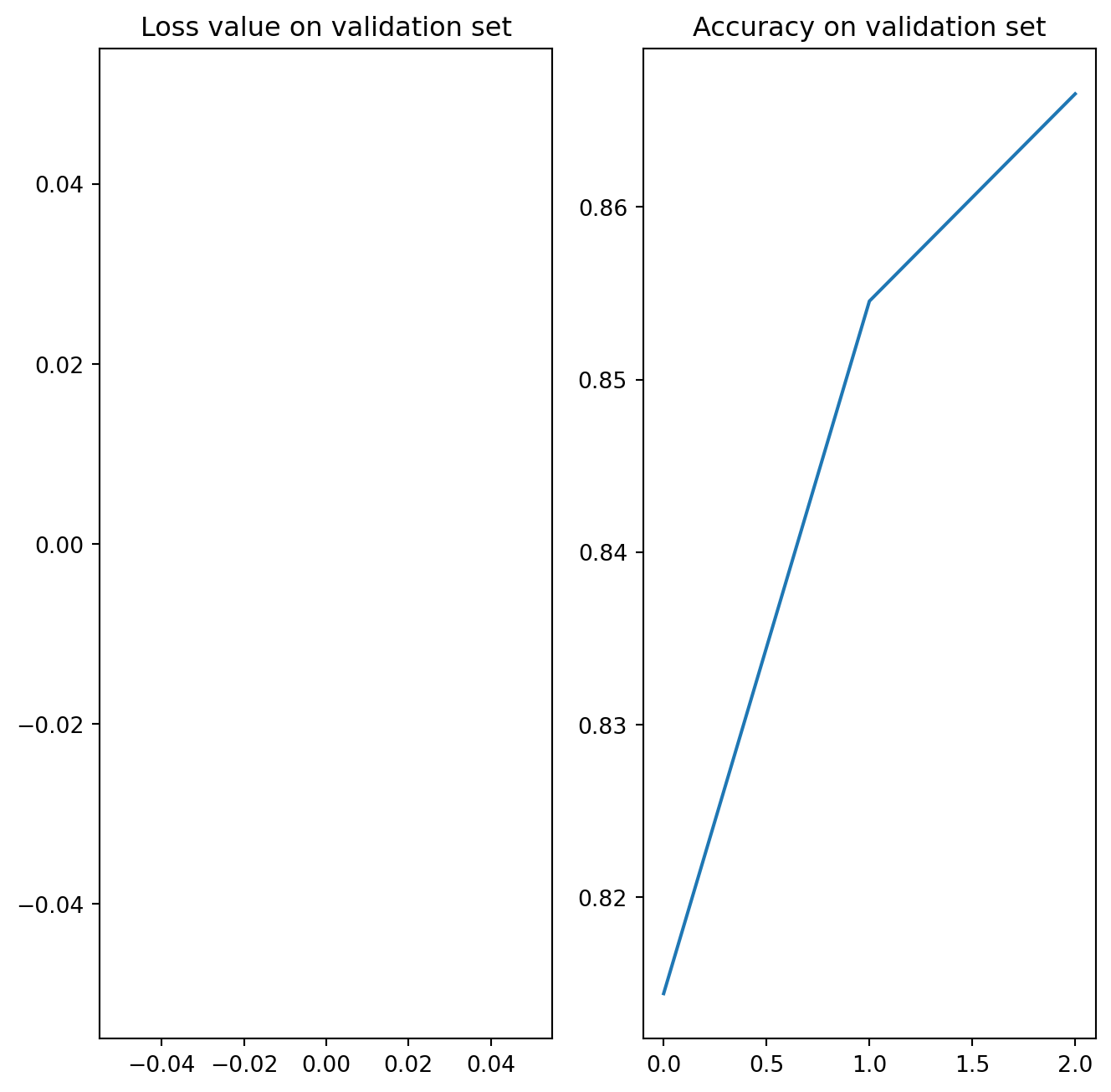
import os
import polars as pl
import imageio.v3 as iio
import grain.python as grain
from jax import random
import dm_pix as pix
import numpy as np
import jax
import jax.numpy as jnp
from flax import nnx
from transformers import FlaxViTForImageClassification
metadata = pl.read_parquet('metadata.parquet')
metadata_train = metadata.filter(pl.col('is_training_img') == 1)
metadata_val = metadata.filter(pl.col('is_training_img') == 0)
cleaned_img_dir = os.path.join(base_dir, 'cleaned_images')
class NABirdsDataset:
"""NABirds dataset class."""
def __init__(self, metadata_file, data_dir):
self.metadata_file = metadata_file
self.data_dir = data_dir
def __len__(self):
return len(self.metadata_file)
def __getitem__(self, idx):
path = os.path.join(self.data_dir, self.metadata_file.get_column('path')[idx])
img = iio.imread(path)
species_name = self.metadata_file.get_column('species_name')[idx]
species_id = self.metadata_file.get_column('species_id')[idx]
photographer = self.metadata_file.get_column('photographer')[idx]
return {
'img': img,
'species_name': species_name,
'species_id': species_id,
'photographer': photographer,
}
nabirds_train = NABirdsDataset(metadata_train, cleaned_img_dir)
nabirds_val = NABirdsDataset(metadata_val, cleaned_img_dir)
class Normalize(grain.MapTransform):
def map(self, element):
img = element['img']
mean = np.array([0.5, 0.5, 0.5], dtype=np.float32)
std = np.array([0.5, 0.5, 0.5], dtype=np.float32)
img = img.astype(np.float32) / 255.0
img_norm = (img - mean) / std
element['img'] = img_norm
return element
class ToFloat(grain.MapTransform):
def map(self, element):
element['img'] = element['img'].astype(np.float32) / 255.0
return element
key = random.key(31)
key, subkey1, subkey2, subkey3, subkey4 = random.split(key, num=5)
class RandomCrop(grain.MapTransform):
def map(self, element):
element['img'] = pix.random_crop(
key=subkey1,
image=element['img'],
crop_sizes=(224, 224, 3)
)
return element
class RandomFlip(grain.MapTransform):
def map(self, element):
element['img'] = pix.random_flip_left_right(
key=subkey2,
image=element['img']
)
return element
class RandomContrast(grain.MapTransform):
def map(self, element):
element['img'] = pix.random_contrast(
key=subkey3,
image=element['img'],
lower=0.8,
upper=1.2
)
return element
class RandomGamma(grain.MapTransform):
def map(self, element):
element['img'] = pix.random_gamma(
key=subkey4,
image=element['img'],
min_gamma=0.6,
max_gamma=1.2
)
return element
class ZScore(grain.MapTransform):
def map(self, element):
img = element['img']
mean = np.array([0.5, 0.5, 0.5], dtype=np.float32)
std = np.array([0.5, 0.5, 0.5], dtype=np.float32)
img = (img - mean) / std
element['img'] = img
return element
seed = 123
train_batch_size = 32
val_batch_size = 2 * train_batch_size
train_sampler = grain.IndexSampler(
num_records=len(nabirds_train),
shuffle=True,
seed=seed,
shard_options=grain.NoSharding(),
num_epochs=1
)
train_loader = grain.DataLoader(
data_source=nabirds_train,
sampler=train_sampler,
operations=[
ToFloat(),
RandomCrop(),
RandomFlip(),
RandomContrast(),
RandomGamma(),
ZScore(),
grain.Batch(train_batch_size, drop_remainder=True)
]
)
val_sampler = grain.IndexSampler(
num_records=len(nabirds_val),
shuffle=False,
seed=seed,
shard_options=grain.NoSharding(),
num_epochs=1
)
val_loader = grain.DataLoader(
data_source=nabirds_val,
sampler=val_sampler,
operations=[
Normalize(),
grain.Batch(val_batch_size)
]
)
class VisionTransformer(nnx.Module):
def __init__(
self,
num_classes: int = 1000,
in_channels: int = 3,
img_size: int = 224,
patch_size: int = 16,
num_layers: int = 12,
num_heads: int = 12,
mlp_dim: int = 3072,
hidden_size: int = 768,
dropout_rate: float = 0.1,
*,
rngs: nnx.Rngs = nnx.Rngs(0),
):
n_patches = (img_size // patch_size) ** 2
self.patch_embeddings = nnx.Conv(
in_channels,
hidden_size,
kernel_size=(patch_size, patch_size),
strides=(patch_size, patch_size),
padding='VALID',
use_bias=True,
rngs=rngs,
)
initializer = jax.nn.initializers.truncated_normal(stddev=0.02)
self.position_embeddings = nnx.Param(
initializer(rngs.params(), (1, n_patches + 1, hidden_size), jnp.float32)
)
self.dropout = nnx.Dropout(dropout_rate, rngs=rngs)
self.cls_token = nnx.Param(jnp.zeros((1, 1, hidden_size)))
self.encoder = nnx.Sequential(*[
TransformerEncoder(hidden_size, mlp_dim, num_heads, dropout_rate, rngs=rngs)
for i in range(num_layers)
])
self.final_norm = nnx.LayerNorm(hidden_size, rngs=rngs)
self.classifier = nnx.Linear(hidden_size, num_classes, rngs=rngs)
def __call__(self, x: jax.Array) -> jax.Array:
patches = self.patch_embeddings(x)
batch_size = patches.shape[0]
patches = patches.reshape(batch_size, -1, patches.shape[-1])
cls_token = jnp.tile(self.cls_token, [batch_size, 1, 1])
x = jnp.concat([cls_token, patches], axis=1)
embeddings = x + self.position_embeddings
embeddings = self.dropout(embeddings)
x = self.encoder(embeddings)
x = self.final_norm(x)
x = x[:, 0]
return self.classifier(x)
class TransformerEncoder(nnx.Module):
def __init__(
self,
hidden_size: int,
mlp_dim: int,
num_heads: int,
dropout_rate: float = 0.0,
*,
rngs: nnx.Rngs = nnx.Rngs(0),
) -> None:
self.norm1 = nnx.LayerNorm(hidden_size, rngs=rngs)
self.attn = nnx.MultiHeadAttention(
num_heads=num_heads,
in_features=hidden_size,
dropout_rate=dropout_rate,
broadcast_dropout=False,
decode=False,
deterministic=False,
rngs=rngs,
)
self.norm2 = nnx.LayerNorm(hidden_size, rngs=rngs)
self.mlp = nnx.Sequential(
nnx.Linear(hidden_size, mlp_dim, rngs=rngs),
nnx.gelu,
nnx.Dropout(dropout_rate, rngs=rngs),
nnx.Linear(mlp_dim, hidden_size, rngs=rngs),
nnx.Dropout(dropout_rate, rngs=rngs),
)
def __call__(self, x: jax.Array) -> jax.Array:
x = x + self.attn(self.norm1(x))
x = x + self.mlp(self.norm2(x))
return x
model = VisionTransformer(num_classes=1000)
tf_model = FlaxViTForImageClassification.from_pretrained('google/vit-base-patch16-224')
def vit_inplace_copy_weights(*, src_model, dst_model):
assert isinstance(src_model, FlaxViTForImageClassification)
assert isinstance(dst_model, VisionTransformer)
tf_model_params = src_model.params
tf_model_params_fstate = nnx.traversals.flatten_mapping(tf_model_params)
flax_model_params = nnx.state(dst_model, nnx.Param)
flax_model_params_fstate = dict(flax_model_params.flat_state())
params_name_mapping = {
('cls_token',): ('vit', 'embeddings', 'cls_token'),
('position_embeddings',): ('vit', 'embeddings', 'position_embeddings'),
**{
('patch_embeddings', x): ('vit', 'embeddings', 'patch_embeddings', 'projection', x)
for x in ['kernel', 'bias']
},
**{
('encoder', 'layers', i, 'attn', y, x): (
'vit', 'encoder', 'layer', str(i), 'attention', 'attention', y, x
)
for x in ['kernel', 'bias']
for y in ['key', 'value', 'query']
for i in range(12)
},
**{
('encoder', 'layers', i, 'attn', 'out', x): (
'vit', 'encoder', 'layer', str(i), 'attention', 'output', 'dense', x
)
for x in ['kernel', 'bias']
for i in range(12)
},
**{
('encoder', 'layers', i, 'mlp', 'layers', y1, x): (
'vit', 'encoder', 'layer', str(i), y2, 'dense', x
)
for x in ['kernel', 'bias']
for y1, y2 in [(0, 'intermediate'), (3, 'output')]
for i in range(12)
},
**{
('encoder', 'layers', i, y1, x): (
'vit', 'encoder', 'layer', str(i), y2, x
)
for x in ['scale', 'bias']
for y1, y2 in [('norm1', 'layernorm_before'), ('norm2', 'layernorm_after')]
for i in range(12)
},
**{
('final_norm', x): ('vit', 'layernorm', x)
for x in ['scale', 'bias']
},
**{
('classifier', x): ('classifier', x)
for x in ['kernel', 'bias']
}
}
nonvisited = set(flax_model_params_fstate.keys())
for key1, key2 in params_name_mapping.items():
assert key1 in flax_model_params_fstate, key1
assert key2 in tf_model_params_fstate, (key1, key2)
nonvisited.remove(key1)
src_value = tf_model_params_fstate[key2]
if key2[-1] == 'kernel' and key2[-2] in ('key', 'value', 'query'):
shape = src_value.shape
src_value = src_value.reshape((shape[0], 12, 64))
if key2[-1] == 'bias' and key2[-2] in ('key', 'value', 'query'):
src_value = src_value.reshape((12, 64))
if key2[-4:] == ('attention', 'output', 'dense', 'kernel'):
shape = src_value.shape
src_value = src_value.reshape((12, 64, shape[-1]))
dst_value = flax_model_params_fstate[key1]
assert src_value.shape == dst_value.value.shape, (key2, src_value.shape, key1, dst_value.value.shape)
dst_value.value = src_value.copy()
assert dst_value.value.mean() == src_value.mean(), (dst_value.value, src_value.mean())
assert len(nonvisited) == 0, nonvisited
# Notice the use of `flax.nnx.update` and `flax.nnx.State`.
nnx.update(dst_model, nnx.State.from_flat_path(flax_model_params_fstate))
vit_inplace_copy_weights(src_model=tf_model, dst_model=model)
model.classifier = nnx.Linear(model.classifier.in_features, 405, rngs=nnx.Rngs(0))